Analysis of an image database with visualea¶
Automatical analysis of a set of root images can be done using an image database. The rhizoscan package provide a visualea dataflow for this task. To open it, doulbe click on arabidopsis pipeline at the bottom of the rhizoscan package:
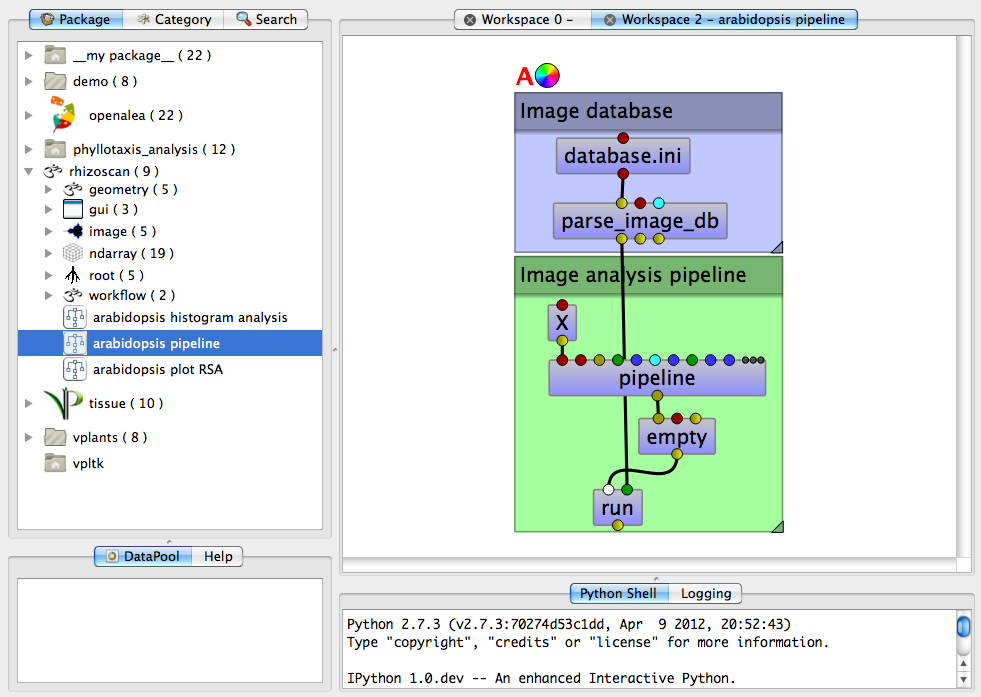
This dataflow is made of two parts:
- The top one loads an image database. It contains 2 modules:
- The first is to indicates the database file to load (see image database for details). By default it points to a little example database contained in the rhizoscan package. If you want toselect another file, double click on the top modules. It opens a file selection user interface where you can browse for the database file you want to load. You will need to have a valid database file in ini file format: see the page on image database for a description.
- The second is the module that load all images from the database. It does not require any configuration.
- The bottom one extracts the root systems from all images. It has two main modules:
- The pipeline module is the image arabidopsis image pipeline which analysis root images.
- The lower module named run is the “start button”: to apply the image pipeline analysis to the whole database, right click on the run module then select run in the menu.